The concept of how to grep search engine results with R had been one of my lingering thoughts. Because sometimes it's just feels easier if I could just grep search engine results than to find a dataset or complete database of a certain thing.
So when at work I had to use ATC codes to get the substances out or the pharmaceutical(?) use. So while no-one was looking I naturally had to devise the following script. This snippet is more or less done, a part of a imaginary whole if you will. It lacks the functionality to search for partial matches. So you can't just look for all the "B01" codes. Probably doing that extension next when I have the time. Also you can't use this for substance to ATC code. But for now this one way ticket, the basic concept as a snippet. Naturally comments about making it more readable and/or efficient are always welcome.
2013-07-20
2013-03-24
A Merging Test Bench
As requested here's the packed data and a test bench you can test your own merging function ideas and replicate my results (hopefully). If you want the plots you can use the end part of scripts in part1 part2.
The data is a bunch of super secret Eve Online killmails. The first part of the script handles the json to data.frame transformation. After that I introduce the R core merging in a for-loop, data.table variant, reduce function variants, and finally the fast.merging. There's actually alot more data then we are using in the bench. Totalling 166106 killmails, merging only 3000 of those. That is because as noted in previous posts the merge for-looping seems to work in exponential time so going much higher then 3000 lines seems really excessive. Even now it took over an hour to run the whole bench on intel i5 3570k (not overclocked). So if you want faster results choose 1000ish frames and set rbenchmark replications to 1. With low frame count (under 100) the results are fairly similiar.
If you are going to test are the results from different functions the same, note that the order of rows and columns will be different. I'm eagerly waiting for you results and ideas.
The data is a bunch of super secret Eve Online killmails. The first part of the script handles the json to data.frame transformation. After that I introduce the R core merging in a for-loop, data.table variant, reduce function variants, and finally the fast.merging. There's actually alot more data then we are using in the bench. Totalling 166106 killmails, merging only 3000 of those. That is because as noted in previous posts the merge for-looping seems to work in exponential time so going much higher then 3000 lines seems really excessive. Even now it took over an hour to run the whole bench on intel i5 3570k (not overclocked). So if you want faster results choose 1000ish frames and set rbenchmark replications to 1. With low frame count (under 100) the results are fairly similiar.
If you are going to test are the results from different functions the same, note that the order of rows and columns will be different. I'm eagerly waiting for you results and ideas.
2013-03-21
RMark: data.table merge vs core merge
This is the third post concerning fast merging in R, first here and second here. This time we are going to look at how the merge function from data.table package works in our case, requested by Uwe Block.
As a reminder the first post concerns doing a merging scheme using the lapply function. After that in the second post we looked how it translates to a parallel solution for even more speed.
As a reminder the problem is the following: given N amount of files with a random amount of variables in each file. How do we merge these files into a complete data within reasonable amount of time? For example you can consider that the medical instruments records it's each run (patient) to a different file. With only the values measured by the instrument are recorded to the file.
In practice I'm using Eve Online killmails. So far I have around 160k mails. Hence the need for a fast solution.
Personally I haven't really used data.table, but it looks good enough. So I recommend checking it out. But in a sense nothing's really changed on my code part. I did a fast merging function based on data.table merge called the lady, but won't give the code because it isn't fast enough (spoiler alert). Also the data.table merge is a generic function. Meaning if you are using data.tables in a merge it automaticaly uses the data.table merge function. But enough it this prattle. Here's the results.
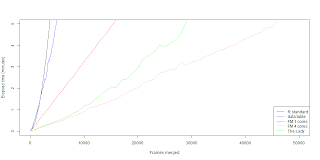
data.table merge is faster then the R core merge function if you are merging a for-loop solution. The problem still is that the data.table merging in a for-loop works in exponential time. Which doesn't translate well for big data. The single core fast merging scheme still beats the data.table merge in a for-loop. The biggest wonder is why when we combine the data.table merge with parallel fast merging scheme we get a slower outcome. Naturally I though we could squeeze few seconds out with low frame count, but seems that it's actually few minutes worse. So far I'm running dry in practical ideas and reasons why the lady seems so fat. Maybe it doesn't cleane up it's diet ? (memory management?) If you have some insight please comment.
As requested, here's the bench.
As a reminder the first post concerns doing a merging scheme using the lapply function. After that in the second post we looked how it translates to a parallel solution for even more speed.
As a reminder the problem is the following: given N amount of files with a random amount of variables in each file. How do we merge these files into a complete data within reasonable amount of time? For example you can consider that the medical instruments records it's each run (patient) to a different file. With only the values measured by the instrument are recorded to the file.
In practice I'm using Eve Online killmails. So far I have around 160k mails. Hence the need for a fast solution.
Personally I haven't really used data.table, but it looks good enough. So I recommend checking it out. But in a sense nothing's really changed on my code part. I did a fast merging function based on data.table merge called the lady, but won't give the code because it isn't fast enough (spoiler alert). Also the data.table merge is a generic function. Meaning if you are using data.tables in a merge it automaticaly uses the data.table merge function. But enough it this prattle. Here's the results.

data.table merge is faster then the R core merge function if you are merging a for-loop solution. The problem still is that the data.table merging in a for-loop works in exponential time. Which doesn't translate well for big data. The single core fast merging scheme still beats the data.table merge in a for-loop. The biggest wonder is why when we combine the data.table merge with parallel fast merging scheme we get a slower outcome. Naturally I though we could squeeze few seconds out with low frame count, but seems that it's actually few minutes worse. So far I'm running dry in practical ideas and reasons why the lady seems so fat. Maybe it doesn't cleane up it's diet ? (memory management?) If you have some insight please comment.
As requested, here's the bench.
2013-03-03
RMark: Parallel fast merging
Last time we looked at a different method of merging multiple files via fast mergin g. I highly recommend checking the fast merging function before we will translate it to a parallel solution. Parallel programming in this case means we are using more then one processor core. So even if you have a fancy 8 core machine R only uses one core if you don't use parallel functions.
In R parallel programming can be done for example with snow package. When we change our function from 1 core solution to a multicore solution we use snow packages parLapply function instead of lapply function. This change will magically make our independent loops run faster without doing any major changes to the script. As parLapply behaves almost exactly the same as lapply. So let us present the script first, the explanation will be given after that. There's alot more case handling in multicore case as we can't call our multicore solution if we only need one core.
For example if we have 100 frames to merge and a 4 core machine. We choose a section size of 25 parts (nparts=25). Which means that we have 4 sections. Section 1 will be frames from 1-25, section 2 will be frames 26-50, and so on. Now we tell the first core to merge the frames from section 1, the second core will merge frames outlined in section 2, and so on. After that is done we only have 4 frames, which can we will merge using the standard way. With this fast merging should atleast two times faster when using 2 cores. Let us see if that is the case, unfortunately I only have 4 cores.
In R parallel programming can be done for example with snow package. When we change our function from 1 core solution to a multicore solution we use snow packages parLapply function instead of lapply function. This change will magically make our independent loops run faster without doing any major changes to the script. As parLapply behaves almost exactly the same as lapply. So let us present the script first, the explanation will be given after that. There's alot more case handling in multicore case as we can't call our multicore solution if we only need one core.
For example if we have 100 frames to merge and a 4 core machine. We choose a section size of 25 parts (nparts=25). Which means that we have 4 sections. Section 1 will be frames from 1-25, section 2 will be frames 26-50, and so on. Now we tell the first core to merge the frames from section 1, the second core will merge frames outlined in section 2, and so on. After that is done we only have 4 frames, which can we will merge using the standard way. With this fast merging should atleast two times faster when using 2 cores. Let us see if that is the case, unfortunately I only have 4 cores.
2013-01-15
Damn syntax highlighting
Well... It seems my syntax highlighting broke... So Github A-HOY! But don't have the time today... Curses...
2013-01-14
R Benchmark: Merge vs fast.merging
Last summer as a trainee I ran into a bit of trouble with the merge function in R. The main problem is that it's slower then your avarage tortoise in a tard field. Also I could not use rbind because of the different lengths of the frames. In code I mean that the next scheme is really slow when you have alot of files:
It wasn't really a problem back in the summer because I didn't have to think it that much. But now it's realy a pain as I need to merge over 160 000 lines. So because I needed to save some time I came up with the following merging algorithm:
The scheme seems completely unintuitive at first. Because you are basically using lapply to vectorize for-loop merging. But it probably makes sense when you have a grasp how vectorizing works in R, or why you should always define the size of your variables before you use them. But enough of that, time for the actual benchmark results:
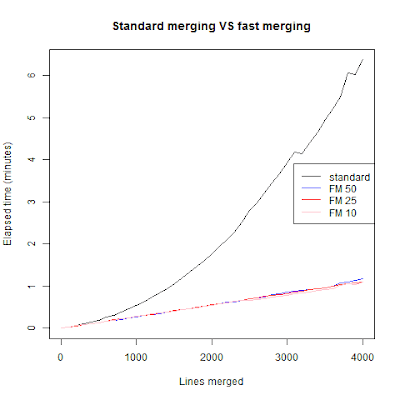
As we can see the standard for-loop merging grows exponentially in time as we merge more frames. The algorithm I used grows in linear time and didn't really depend on the nparts argument. Meaning that with nparts being 50, 25 or 10 the time difference is few seconds when we are at 4001 merges. I didn't want to go further because that for-looping get's really tedious to wait. I think we can do better then fast merging, for one we can use parallel calculations. But that's for next week (read: hopefully this weekend).
Benchmark was created with this scheme:
complete = read.csv(yourfiles[1])
for(i in 2:length(yourfiles)){
part = read.csv(yourfiles[i])
complete = merge(complete, part, all=TRUE, sort=FALSE)
}
It wasn't really a problem back in the summer because I didn't have to think it that much. But now it's realy a pain as I need to merge over 160 000 lines. So because I needed to save some time I came up with the following merging algorithm:
#data.list = list of your frames you want to merge
#nparts = how many parts are merged inside apply, the optimal value is probably
#different for every machine, but should not be a big deal.
fast.merging = function(data.list, nparts=10){
if(!is.list(data.list)) stop("data.list isn't a list") #standard error
while(length(data.list) != 1){ #Loop until everything is merged
#define sections according to nparts
if(length(data.list) > nparts){
starts = seq(1, length(data.list), nparts)
ends = seq(nparts, length(data.list), nparts)
#starts and ends are of equal size
#if length(data.list) divides nparts.
if(length(ends) < length(starts)) ends = c(ends, length(data.list))
#making sure things are even
sections = matrix(c(starts, ends), ncol=2, byrow=FALSE)
sections = apply(sections, 1, list)
}else{
sections = list(c(1, length(data.list)))
}
#We have the standard way inside lapply
#lapply merges a length(sections) amount of nparts sections
data.list = lapply(sections, function(x, data.list){
if(is.list(x)) x = x[[1]]
#the standard way starts ->
part = data.list[[x[1]]]
for(i in x[1]:x[2]){
part = merge(part, data.list[[i]], all=TRUE, sort=FALSE)
}
#<- standard way ends
return(part)
}, data.list = data.list)
#rinse and repeat until all is done
}
return(data.list[[1]]) #returning the merged data frame
}
The scheme seems completely unintuitive at first. Because you are basically using lapply to vectorize for-loop merging. But it probably makes sense when you have a grasp how vectorizing works in R, or why you should always define the size of your variables before you use them. But enough of that, time for the actual benchmark results:

As we can see the standard for-loop merging grows exponentially in time as we merge more frames. The algorithm I used grows in linear time and didn't really depend on the nparts argument. Meaning that with nparts being 50, 25 or 10 the time difference is few seconds when we are at 4001 merges. I didn't want to go further because that for-looping get's really tedious to wait. I think we can do better then fast merging, for one we can use parallel calculations. But that's for next week (read: hopefully this weekend).
Benchmark was created with this scheme:
#Getting our lines to a list of lines
#my frames, in this case only line per data-frame.
eve.frames = lapply(eve.list, eve.data.frame) #Some Eve Online kill mails
sekvenssi = seq(1, 4001, 100) #only trying 1, 101, 201, ... 4001.
time1 = rep(NA, length(sekvenssi))
for(i in 1:length(sekvenssi)){
complete = eve.frames[[1]]
time1[i] = system.time(
for(j in 2:sekvenssi[i]){
complete = merge(complete, eve.frames[[j]],
all=TRUE, sort=FALSE)
}
)[3]
print(paste(round(time1[i],2), nrow(complete),i))
}
write(time1, "time1.dat")
#backup = complete
time2 = rep(NA, length(sekvenssi))
for(i in seq(sekvenssi)){
complete = eve.frames[1:sekvenssi[i]]
time2[i] = system.time(fast.merging(complete, 50))[3]
print(paste(round(time2[i],2), nrow(complete),i))
}
write(time2, "time2.dat")
time3 = rep(NA, length(sekvenssi))
for(i in seq(sekvenssi)){
complete = eve.frames[1:sekvenssi[i]]
time3[i] = system.time(fast.merging(complete, 25))[3]
print(paste(round(time3[i],2), nrow(complete),i))
}
write(time3, "time3.dat")
time4 = rep(NA, length(sekvenssi))
for(i in seq(sekvenssi)){
complete = eve.frames[1:sekvenssi[i]]
time4[i] = system.time(fast.merging(complete, 10))[3]
print(paste(round(time4[i],2), nrow(complete),i))
}
write(time4, "time4.dat")
#To check if they are correct try
#complete = fast.merging(complete, 10)
#backup = backup[order(backup[, 1]), order(colnames(backup))]
#complete = complete[order(complete[, 1]), order(colnames(complete))]
#sum(backup == complete, na.rm=TRUE)
#sum(is.na(backup))
#these should add up to the matrix size.
plot(sekvenssi, time1/60, type="l", ylab="Elapsed time (minutes)", xlab="Lines merged", main="Standard merging VS fast merging")
points(sekvenssi, time2/60, type="l", col="blue")
points(sekvenssi, time3/60, type="l", col="red")
points(sekvenssi, time4/60, type="l", col="pink")
legend("right", c("standard", "FM 50", "FM 25", "FM 10"),
lty=c(1,1,1,1), col=c("black", "blue", "red", "pink"))
2013-01-07
Comment: Search and Replace
A post in R bloggers caught my attention this morning. The main idea was that how can you change objects in a string. For example given a basket of fruits we would like to change apples to bananas by using R and the ifelse funtion. There are two main solutions how to change one object into another:
If you want to change multiple objects you would need an equal amount of ifelses. In the original post, here, R de jeu presented his function solution which removes the cumbersome writing. I recommend reading the orginal post if the idea isn't immediately clear.
The function given in the post by R de jeu is the following
I'm not a big fan of recursive functions as they tend to be the slowest of all possible solutions. So I decided to do my own version of the decode function without recursion.
A standard for-looping with extra logical operations. Now it's time for some speed tests! Spoilers: my solution was around 20ish times faster, but with small vectors the time difference isn't that important.
#Given a basket of fruits
basket = c("apple", "banana", "lemon", "orange", "orange", "pear", "cherry")
basket = ifelse(basket == "apple", "banana" , basket)
basket[basket == "lemon"] = "pear"
#The latter is usually preferred, but sometimes the ifelse cannot be avoided
If you want to change multiple objects you would need an equal amount of ifelses. In the original post, here, R de jeu presented his function solution which removes the cumbersome writing. I recommend reading the orginal post if the idea isn't immediately clear.
The function given in the post by R de jeu is the following
decode <- function(x, search, replace, default = NULL) {
# build a nested ifelse function by recursion
decode.fun <- function(search, replace, default = NULL)
if (length(search) == 0L) {
function(x) if (is.null(default)) x else rep(default, length(x))
} else {
function(x) ifelse(x == search[1L], replace[1L],
decode.fun(tail(search, -1L),
tail(replace, -1L),
default)(x))
}
return(decode.fun(search, replace, default)(x))
}
I'm not a big fan of recursive functions as they tend to be the slowest of all possible solutions. So I decided to do my own version of the decode function without recursion.
#Same input as in the decode function
decode2 = function(x, search, replace, default=NULL){
#this is if we want the "default type" output
if(!is.null(default)){
default.bol = !(x %in% search)
x[default.bol] = default
}
for(i in seq(search)){
x[x == search[i]] = replace[i]
}
return(x)
}
A standard for-looping with extra logical operations. Now it's time for some speed tests! Spoilers: my solution was around 20ish times faster, but with small vectors the time difference isn't that important.
#If it's worth doing, it's worth overdoing
picnic.basket <- c("apple", "banana", "Lightsaber", "pineapple", "strawberry",
"lemon","joy","orange", "genelec speakers", "cat", "pear", "cherry",
"beer", "sonic screwdriver", "cat with a caption", "knife", "cheddar",
"The Book of Proofs", "evil ladies", "cheese", "The R inferno", "smoked reindeer meat",
"salmon", "Tardis", "Book of Death", "Pillows", "Blanket", "Woman")
multi.dimensional.basket = sample(picnic.basket, 1000000, replace=TRUE)
search = sample(picnic.basket, 10)
replace = sample(picnic.basket[!(picnic.basket %in% search)], 10)
#Making sure results match.
sum(decode(multi.dimensional.basket, search, replace) ==
decode2(multi.dimensional.basket, search, replace))
sum(decode(multi.dimensional.basket, search, replace, "fig") ==
decode2(multi.dimensional.basket, search, replace, "fig"))
system.time( decode(multi.dimensional.basket, search, replace) )
system.time( decode2(multi.dimensional.basket, search, replace) )
system.time( decode(multi.dimensional.basket, search, replace, "fig") )
system.time( decode2(multi.dimensional.basket, search, replace, "fig") )
2013-01-05
National identification number: Finland part 3
Last part of our short series about the Finnish social security number (Fssn). You can check part 1 here, and part 2 here.
This last post we are interested in generating random Fssn's. This has no real world applications. It is just a coding excercise for the willing. If you are up for the challenge try doing it yourself.
I'm not going to go indepth about this algorithm, I'll just present it straight away.
This last post we are interested in generating random Fssn's. This has no real world applications. It is just a coding excercise for the willing. If you are up for the challenge try doing it yourself.
I'm not going to go indepth about this algorithm, I'll just present it straight away.
#n the amount of random generated FSSn
#pos.centuries are the possible centuries you want to generate
#pos.century.char are the characters which match the possible centuries
#pos.centuries and pos.century.char have to be in the same order.
finID.random = function(n, pos.centuries = c(18, 19, 20), pos.century.char=c("+", "-", "A")){
# this function is for genereting one random Finnish ID. It's later used in an apply function.
#A function inside a function! Smart...
finID.randomizer = function(x, centuries, century.char){
# x is just a dummy variable so that apply works.
cent.char = sample(century.char, 1)
cent = centuries[century.char %in% cent.char]
year = sample(1:99, 1)
if(nchar(year) ==1) year = paste("0", year, sep="")
bol.year = as.integer(paste(cent, year, sep=""))
month = sample(1:12, 1)
#Leap year handling... Straight from wikipedia
if(bol.year %% 400 == 0){
days = c(31, 29, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31)[month]
}else if(bol.year %% 100 == 0){
days = c(31, 28, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31)[month]
}else if(bol.year %% 4 == 0){
days = c(31, 29, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31)[month]
}else{
days = c(31, 28, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31)[month]
}
days = sample(1:days, 1)
if(nchar(month) == 1) month = paste("0", month, sep="")
if(nchar(days) == 1) days = paste("0", days, sep="")
birth = paste(days, month, year, sep="")
check.value = sample(0:999, 1)
if(nchar(check.value) == 3) {
}else if(nchar(check.value) == 2){
check.value = paste("0", check.value, sep="")
}else{
check.value = paste("00", check.value, sep="")
}
check.char = c(0, 1, 2, 3, 4, 5, 6, 7, 8, 9, LETTERS)
check.char = check.char[-c(17, 19, 25, 27, 36)]
bol.value = as.integer(paste(birth, check.value, sep=""))
check.char = check.char[(bol.value %% 31) + 1]
x = paste(birth, cent.char, check.value, check.char, sep="")
}
#formals "reformats" a given function
formals(finID.randomizer) = alist(x=,
centuries = pos.centuries,
century.char = pos.century.char)
n = matrix(NA, nrow = n)
n = apply(n, 1, finID.randomizer)
return(n)
}
National identification number: Finland part2
Continuing our theme from last time. The Finnish social security number (FSSn) has the form xxxxxxyzzzq, where the check digit is q.
If you want to check if the FSSn is real the check digit is matched to the remainder of xxxxxxzzz / 31. The check digit can be numbers from 0-9 followed by letters A, B, C, D, E, F, H, J, K, L, M, N, P, Q, R, S, T, U, V, W, X, Y. If the remainder is a number between 0-9 then it's matched to the numbers 0-9, if the remainder is from 10-30 it's matched to the letters in the order they are given. So for example 10 is A and 20 is M.
It makes sense to drop O, I, and Z from the alphabet because they can be confused to 0, 1 and 2, but quite often handwritten S and 5 get mixed up.
The algorithm in R would be the following
If you want to check if the FSSn is real the check digit is matched to the remainder of xxxxxxzzz / 31. The check digit can be numbers from 0-9 followed by letters A, B, C, D, E, F, H, J, K, L, M, N, P, Q, R, S, T, U, V, W, X, Y. If the remainder is a number between 0-9 then it's matched to the numbers 0-9, if the remainder is from 10-30 it's matched to the letters in the order they are given. So for example 10 is A and 20 is M.
It makes sense to drop O, I, and Z from the alphabet because they can be confused to 0, 1 and 2, but quite often handwritten S and 5 get mixed up.
The algorithm in R would be the following
#Given x a vector of FSSn's
finID.real = function(x){
#if the amount of characters of a certain FSSn is not 11
#or some FSSn is NA we change them as ""
if(sum(nchar(x) != 11 | is.na(x)) >= 1) FSSn[nchar(x) != 11 | is.na(x) >= 1] = ""
check.char = c(0,1,2,3,4,5,6,7,8,9,LETTERS) #LETTERS equals the whole alphabet
check.char = check.char[-c(17,19,25,27,36)] #We remove the unwated letters
last.chars = substr(x,11,11) #The check digits
x = matrix(x,nrow=length(x)) #The standard trick so that apply works.
x = apply(x, 1, function(x) {
# as.integer("5") gives out 5.
x = as.integer(paste(substr(x,1,6),substr(x,8,10),sep="")) %% 31
x = x+1 #We add +1 because vectors in R begin from 1 not 0.
return(x)
}
)
#the ifelse return either the check.char or NA
#which is then matched to the last character
bol = last.chars == ifelse(!is.na(x), check.char[x], NA)
if(sum(is.na(bol)) >= 1) bol[is.na(bol)] = FALSE #Chancing NAs to FALSE
return(bol) #Returning a TRUE / FALSE vector.
}
Subscribe to:
Posts (Atom)